
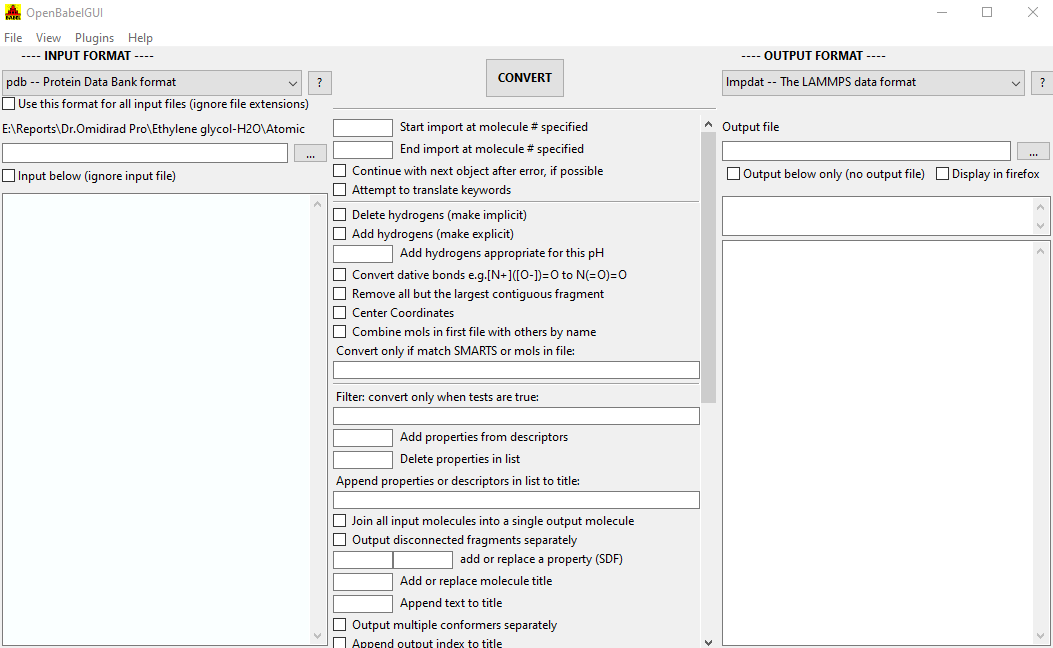
See -H format-ID for options allowed 25 by a particular format 26 27 -addtotitle 28 Append text to the current molecule title 29 30 -addformula 31 Append the molecular formula after the current molecule title 32 33 -b Convert dative bonds: e.g., ()=O to N(=O)=O 34 35 -c Center atomic coordinates at (0,0,0) 36 37 -C Combine molecules in first file with others having the same name 38 39 -e Continue after errors 40 41 -d Delete Hydrogens 42 43 -errorlevel 2 44 Filter the level of errors and warnings displayed: 45 1 = critical errors only 46 2 = include warnings too (default) 47 3 = include informational messages too 48 4 = include "audit log" messages of changes to data 49 5 = include debugging messages too 50 51 -f # For multiple entry input, start import with molecule # as the first 52 entry 53 54 -F Output the available fingerprint types 55 56 -h Add hydrogens 57 58 -H Output usage information 59 60 -H format-ID 61 Output formatting information and options for format specified 62 63 -Hall 64 Output formatting information and options for all formats 65 66 -i 67 Specifies input format, see below for the available formats 68 69 -j 70 71 -join 72 Join all input molecules into a single output molecule entry 73 74 -k Translate computational chemistry modeling keywords (e.g., GAMESS 75 and Gaussian) 76 77 -m Produce multiple output files, to allow: 78 - Splitting one input file - put each molecule into consec‐ 79 utively numbered output files 80 - Batch conversion - convert each of multiple input files 81 into a specified output format 82 83 -l # For multiple entry input, stop import with molecule # as the last 84 entry 85 86 -o format-ID 87 Specifies output format, see below for the available formats 88 89 -p Add Hydrogens appropriate for pH (use transforms in phmodel.txt) 90 91 -property 92 Add or replace a property (e.g., in an MDL SD file) 93 94 -s SMARTS 95 Convert only molecules matching the SMARTS pattern specified 96 97 -separate 98 Separate disconnected fragments into individual molecular records 99 100 -t All input files describe a single molecule 101 102 -title title 103 Add or replace molecular title 104 105 -x options 106 Format-specific output options. 22 23 -a options 24 Format-specific input options. 18 OPTIONS 20 If only input and ouput files are given, Open Babel will guess the file 21 type from the filename extension. For more information, se the Open Babel web pages 17.
14 15 Open Babel is also a complete programmers toolkit for developing chem‐ 16 istry software.

Openbabel zmatrix output manual#
1 babel(1) User's Reference Manual babel(1) 2 NAME 4 babel - a converter for chemistry and molecular modeling data files 5 SYNOPSIS 7 babel 8 babel infile outfile 9 DESCRIPTION 11 Babel is a cross-platform program designed to interconvert between many 12 file formats used in molecular modeling and computational chemistry and 13 related areas.


 0 kommentar(er)
0 kommentar(er)
